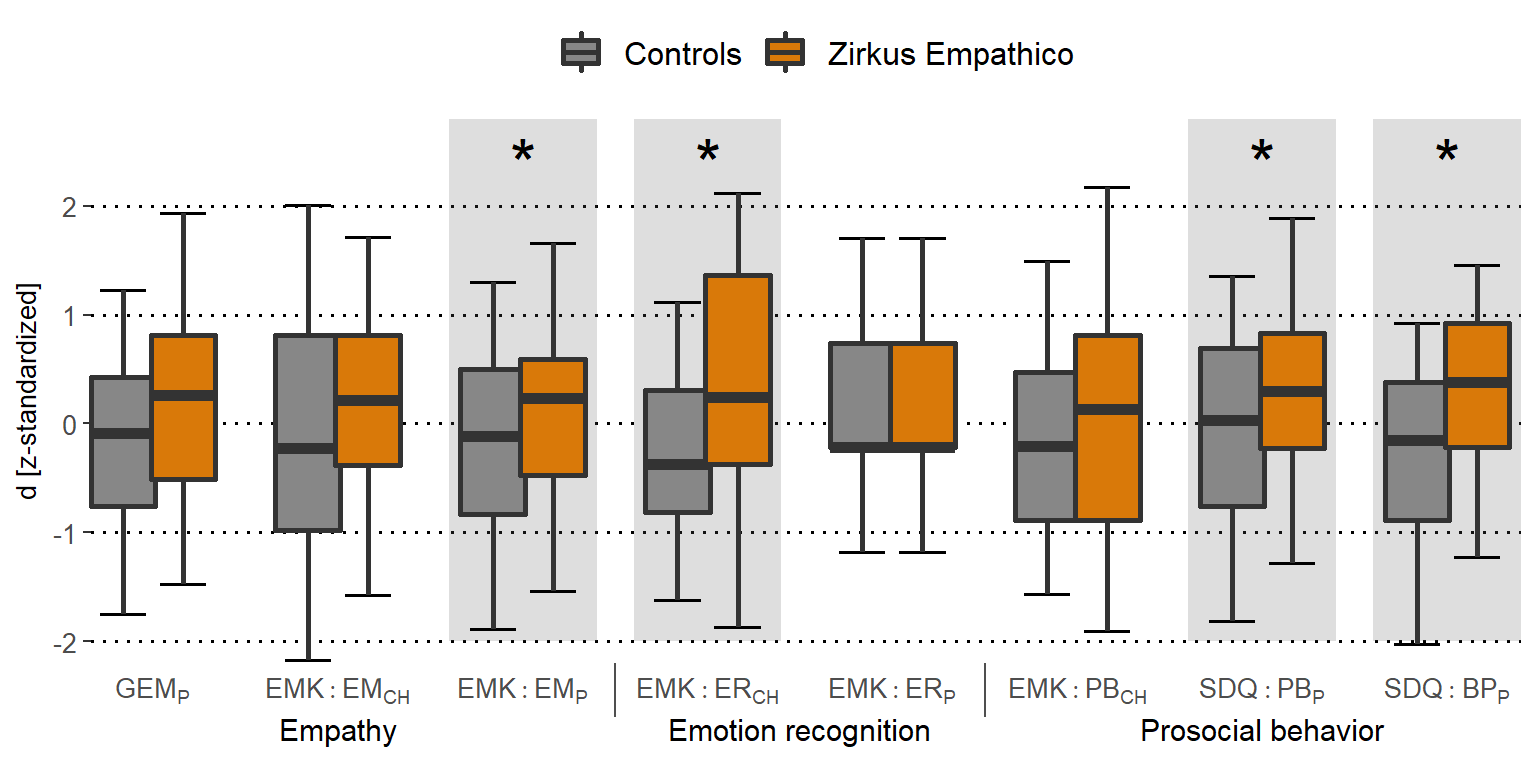
Primary outcome: Empathy
GEM
ITT
Without training time
gem_total_an = aov_ez("ID", "d_GEM_Total", qn_data_itt, between = c("group"), covariate = c("GEM_Total_T1"),
observed = c("GEM_Total_T1"), factorize = FALSE, anova_table = list(correction = "none",
es = "pes"))
pander(gem_total_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 71 | 184.2 | 0.9933 | 0.0138 | 0.3223 |
| GEM_Total_T1 | 1 | 71 | 184.2 | 6.774 | 0.0871 | 0.01125 |
# https://thenewstatistics.com/itns/2020/07/04/3-easy-ways-to-obtain-cohens-d-and-its-ci/
# Calculate mean and SD (and square them) for T2-T1 difference per group (ZE and
# CG)
mean_rel_CG = mean(qn_data_itt[qn_data_itt$group == "CG", ]$d_GEM_Total)
mean_rel_TG = mean(qn_data_itt[qn_data_itt$group == "TG", ]$d_GEM_Total)
sd_rel_CG = sd(qn_data_itt[qn_data_itt$group == "CG", ]$d_GEM_Total)
sd_rel_TG = sd(qn_data_itt[qn_data_itt$group == "TG", ]$d_GEM_Total)
# Calculate difference between TG and CG
diff_mean_TG_CG = mean_rel_TG - mean_rel_CG
# Get number of participants per group
pnum = table(qn_data_itt["group"])
pnum_CG = pnum[1]
pnum_TG = pnum[2]
# Get Cohen's and confidence intervals
estimate = estimateStandardizedMeanDifference(mean_rel_TG, mean_rel_CG, sd_rel_TG,
sd_rel_CG, pnum_TG, pnum_CG, conf.level = 0.95)
# Retrieve them from list
GEM_Total_d_itt = estimate$cohend
GEM_Total_low_d_itt = estimate$cohend.low
GEM_Total_high_d_itt = estimate$cohend.highWith training time
gem_total_an = aov_ez("ID", "d_GEM_Total", qn_data_itt, between = c("group"), covariate = c("GEM_Total_T1",
"train_time"), observed = c("GEM_Total_T1", "train_time"), factorize = FALSE,
anova_table = list(correction = "none", es = "pes"))
pander(gem_total_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 67 | 183.9 | 0.3401 | 0.00505 | 0.5617 |
| GEM_Total_T1 | 1 | 67 | 183.9 | 4.909 | 0.06826 | 0.03012 |
| train_time | 1 | 67 | 183.9 | 2.111 | 0.03055 | 0.1509 |
PP
Without training time
gem_total_an = aov_ez("ID", "d_GEM_Total", qn_data_pp, between = c("group"), covariate = c("GEM_Total_T1"),
observed = c("GEM_Total_T1"), factorize = FALSE, anova_table = list(correction = "none",
es = "pes"))
pander(gem_total_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 69 | 182.7 | 0.8724 | 0.01249 | 0.3535 |
| GEM_Total_T1 | 1 | 69 | 182.7 | 5.191 | 0.06997 | 0.0258 |
# https://thenewstatistics.com/itns/2020/07/04/3-easy-ways-to-obtain-cohens-d-and-its-ci/
# Calculate mean and SD (and square them) for T2-T1 difference per group (ZE and
# CG)
mean_rel_CG = mean(qn_data_pp[qn_data_pp$group == "CG", ]$d_GEM_Total)
mean_rel_TG = mean(qn_data_pp[qn_data_pp$group == "TG", ]$d_GEM_Total)
sd_rel_CG = sd(qn_data_pp[qn_data_pp$group == "CG", ]$d_GEM_Total)
sd_rel_TG = sd(qn_data_pp[qn_data_pp$group == "TG", ]$d_GEM_Total)
# Calculate difference between TG and CG
diff_mean_TG_CG = mean_rel_TG - mean_rel_CG
# Get number of participants per group
pnum = table(qn_data_pp["group"])
pnum_CG = pnum[1]
pnum_TG = pnum[2]
# Get Cohen's and confidence intervals
estimate = estimateStandardizedMeanDifference(mean_rel_TG, mean_rel_CG, sd_rel_TG,
sd_rel_CG, pnum_TG, pnum_CG, conf.level = 0.95)
# Retrieve them from list
GEM_Total_d_pp = estimate$cohend
GEM_Total_low_d_pp = estimate$cohend.low
GEM_Total_high_d_pp = estimate$cohend.highWith training time
gem_total_an = aov_ez("ID", "d_GEM_Total", qn_data_pp, between = c("group"), covariate = c("GEM_Total_T1",
"train_time"), observed = c("GEM_Total_T1", "train_time"), factorize = FALSE,
anova_table = list(correction = "none", es = "pes"))
pander(gem_total_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 66 | 184.6 | 0.4706 | 0.00708 | 0.4951 |
| GEM_Total_T1 | 1 | 66 | 184.6 | 5.123 | 0.07203 | 0.02691 |
| train_time | 1 | 66 | 184.6 | 2.125 | 0.03119 | 0.1497 |
EMK 3-6 EM CH
ITT
Without training time
# ANCOVA EMK_EM_CH
EMK_EM_CH_an = aov_ez("ID", "d_EMK_EM_CH", qn_data_itt, between = c("group"), covariate = c("EMK_EM_CH_T1"),
observed = c("EMK_EM_CH_T1"), factorize = FALSE, anova_table = list(correction = "none",
es = "pes"))
pander(EMK_EM_CH_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 71 | 9.256 | 3.498 | 0.04695 | 0.06558 |
| EMK_EM_CH_T1 | 1 | 71 | 9.256 | 15.49 | 0.1791 | 0.0001916 |
# https://thenewstatistics.com/itns/2020/07/04/3-easy-ways-to-obtain-cohens-d-and-its-ci/
# Calculate mean and SD (and square them) for T2-T1 difference per group (ZE and
# CG)
mean_rel_CG = mean(qn_data_itt[qn_data_itt$group == "CG", ]$d_EMK_EM_CH)
mean_rel_TG = mean(qn_data_itt[qn_data_itt$group == "TG", ]$d_EMK_EM_CH)
sd_rel_CG = sd(qn_data_itt[qn_data_itt$group == "CG", ]$d_EMK_EM_CH)
sd_rel_TG = sd(qn_data_itt[qn_data_itt$group == "TG", ]$d_EMK_EM_CH)
# Calculate difference between TG and CG
diff_mean_TG_CG = mean_rel_TG - mean_rel_CG
# Get number of participants per group
pnum = table(qn_data_itt["group"])
pnum_CG = pnum[1]
pnum_TG = pnum[2]
# Get Cohen's and confidence intervals
estimate = estimateStandardizedMeanDifference(mean_rel_TG, mean_rel_CG, sd_rel_TG,
sd_rel_CG, pnum_TG, pnum_CG, conf.level = 0.95)
# Retrieve them from list
EMK_EM_CH_d_itt = estimate$cohend
EMK_EM_CH_low_d_itt = estimate$cohend.low
EMK_EM_CH_high_d_itt = estimate$cohend.highWith training time
# ANCOVA EMK_EM_CH
EMK_EM_CH_an = aov_ez("ID", "d_EMK_EM_CH", qn_data_itt, between = c("group"), covariate = c("EMK_EM_CH_T1",
"train_time"), observed = c("EMK_EM_CH_T1", "train_time"), factorize = FALSE,
anova_table = list(correction = "none", es = "pes"))
pander(EMK_EM_CH_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 67 | 9.185 | 3.161 | 0.04506 | 0.07993 |
| EMK_EM_CH_T1 | 1 | 67 | 9.185 | 16.67 | 0.1993 | 0.0001207 |
| train_time | 1 | 67 | 9.185 | 0.6096 | 0.009016 | 0.4377 |
PP
Without training time
# ANCOVA EMK_EM_CH
EMK_EM_CH_an = aov_ez("ID", "d_EMK_EM_CH", qn_data_pp, between = c("group"), covariate = c("EMK_EM_CH_T1"),
observed = c("EMK_EM_CH_T1"), factorize = FALSE, anova_table = list(correction = "none",
es = "pes"))
pander(EMK_EM_CH_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 69 | 9.306 | 2.917 | 0.04057 | 0.09213 |
| EMK_EM_CH_T1 | 1 | 69 | 9.306 | 16.42 | 0.1922 | 0.0001313 |
# https://thenewstatistics.com/itns/2020/07/04/3-easy-ways-to-obtain-cohens-d-and-its-ci/
# Calculate mean and SD (and square them) for T2-T1 difference per group (ZE and
# CG)
mean_rel_CG = mean(qn_data_pp[qn_data_pp$group == "CG", ]$d_EMK_EM_CH)
mean_rel_TG = mean(qn_data_pp[qn_data_pp$group == "TG", ]$d_EMK_EM_CH)
sd_rel_CG = sd(qn_data_pp[qn_data_pp$group == "CG", ]$d_EMK_EM_CH)
sd_rel_TG = sd(qn_data_pp[qn_data_pp$group == "TG", ]$d_EMK_EM_CH)
# Calculate difference between TG and CG
diff_mean_TG_CG = mean_rel_TG - mean_rel_CG
# Get number of participants per group
pnum = table(qn_data_pp["group"])
pnum_CG = pnum[1]
pnum_TG = pnum[2]
# Get Cohen's and confidence intervals
estimate = estimateStandardizedMeanDifference(mean_rel_TG, mean_rel_CG, sd_rel_TG,
sd_rel_CG, pnum_TG, pnum_CG, conf.level = 0.95)
# Retrieve them from list
EMK_EM_CH_d_pp = estimate$cohend
EMK_EM_CH_low_d_pp = estimate$cohend.low
EMK_EM_CH_high_d_pp = estimate$cohend.highWith training time
# ANCOVA EMK_EM_CH
EMK_EM_CH_an = aov_ez("ID", "d_EMK_EM_CH", qn_data_pp, between = c("group"), covariate = c("EMK_EM_CH_T1",
"train_time"), observed = c("EMK_EM_CH_T1", "train_time"), factorize = FALSE,
anova_table = list(correction = "none", es = "pes"))
pander(EMK_EM_CH_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 66 | 9.324 | 3.07 | 0.04445 | 0.08439 |
| EMK_EM_CH_T1 | 1 | 66 | 9.324 | 16.35 | 0.1986 | 0.00014 |
| train_time | 1 | 66 | 9.324 | 0.6005 | 0.009016 | 0.4412 |
EMK 3-6 EM P
ITT
Without training time
EMK_EM_P_an = aov_ez("ID", "d_EMK_EM_P", qn_data_itt, between = c("group"), covariate = c("EMK_EM_P_T1"),
observed = c("EMK_EM_P_T1"), factorize = FALSE, anova_table = list(correction = "none",
es = "pes"))
pander(EMK_EM_P_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 71 | 6.603 | 4.136 | 0.05505 | 0.04571 |
| EMK_EM_P_T1 | 1 | 71 | 6.603 | 15.11 | 0.1755 | 0.0002253 |
# https://thenewstatistics.com/itns/2020/07/04/3-easy-ways-to-obtain-cohens-d-and-its-ci/
# Calculate mean and SD (and square them) for T2-T1 difference per group (ZE and
# CG)
mean_rel_CG = mean(qn_data_itt[qn_data_itt$group == "CG", ]$d_EMK_EM_P)
mean_rel_TG = mean(qn_data_itt[qn_data_itt$group == "TG", ]$d_EMK_EM_P)
sd_rel_CG = sd(qn_data_itt[qn_data_itt$group == "CG", ]$d_EMK_EM_P)
sd_rel_TG = sd(qn_data_itt[qn_data_itt$group == "TG", ]$d_EMK_EM_P)
# Calculate difference between TG and CG
diff_mean_TG_CG = mean_rel_TG - mean_rel_CG
# Get number of participants per group
pnum = table(qn_data_itt["group"])
pnum_CG = pnum[1]
pnum_TG = pnum[2]
# Get Cohen's and confidence intervals
estimate = estimateStandardizedMeanDifference(mean_rel_TG, mean_rel_CG, sd_rel_TG,
sd_rel_CG, pnum_TG, pnum_CG, conf.level = 0.95)
# Retrieve them from list
EMK_EM_P_d_itt = estimate$cohend
EMK_EM_P_low_d_itt = estimate$cohend.low
EMK_EM_P_high_d_itt = estimate$cohend.highWith training time
EMK_EM_P_an = aov_ez("ID", "d_EMK_EM_P", qn_data_itt, between = c("group"), covariate = c("EMK_EM_P_T1",
"train_time"), observed = c("EMK_EM_P_T1", "train_time"), factorize = FALSE,
anova_table = list(correction = "none", es = "pes"))
pander(EMK_EM_P_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 67 | 6.472 | 5.601 | 0.07714 | 0.02085 |
| EMK_EM_P_T1 | 1 | 67 | 6.472 | 16.64 | 0.199 | 0.0001224 |
| train_time | 1 | 67 | 6.472 | 0.07457 | 0.001112 | 0.7856 |
PP
Without training time
EMK_EM_P_an = aov_ez("ID", "d_EMK_EM_P", qn_data_pp, between = c("group"), covariate = c("EMK_EM_P_T1"),
observed = c("EMK_EM_P_T1"), factorize = FALSE, anova_table = list(correction = "none",
es = "pes"))
pander(EMK_EM_P_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 69 | 6.377 | 6.004 | 0.08005 | 0.01681 |
| EMK_EM_P_T1 | 1 | 69 | 6.377 | 19.15 | 0.2172 | 0.00004194 |
# https://thenewstatistics.com/itns/2020/07/04/3-easy-ways-to-obtain-cohens-d-and-its-ci/
# Calculate mean and SD (and square them) for T2-T1 difference per group (ZE and
# CG)
mean_rel_CG = mean(qn_data_pp[qn_data_pp$group == "CG", ]$d_EMK_EM_P)
mean_rel_TG = mean(qn_data_pp[qn_data_pp$group == "TG", ]$d_EMK_EM_P)
sd_rel_CG = sd(qn_data_pp[qn_data_pp$group == "CG", ]$d_EMK_EM_P)
sd_rel_TG = sd(qn_data_pp[qn_data_pp$group == "TG", ]$d_EMK_EM_P)
# Calculate difference between TG and CG
diff_mean_TG_CG = mean_rel_TG - mean_rel_CG
# Get number of participants per group
pnum = table(qn_data_pp["group"])
pnum_CG = pnum[1]
pnum_TG = pnum[2]
# Get Cohen's and confidence intervals
estimate = estimateStandardizedMeanDifference(mean_rel_TG, mean_rel_CG, sd_rel_TG,
sd_rel_CG, pnum_TG, pnum_CG, conf.level = 0.95)
# Retrieve them from list
EMK_EM_P_d_pp = estimate$cohend
EMK_EM_P_low_d_pp = estimate$cohend.low
EMK_EM_P_high_d_pp = estimate$cohend.highWith training time
EMK_EM_P_an = aov_ez("ID", "d_EMK_EM_P", qn_data_pp, between = c("group"), covariate = c("EMK_EM_P_T1",
"train_time"), observed = c("EMK_EM_P_T1", "train_time"), factorize = FALSE,
anova_table = list(correction = "none", es = "pes"))
pander(EMK_EM_P_an$anova_table)| num Df | den Df | MSE | F | pes | Pr(>F) | |
|---|---|---|---|---|---|---|
| group | 1 | 66 | 6.55 | 5.694 | 0.07942 | 0.0199 |
| EMK_EM_P_T1 | 1 | 66 | 6.55 | 16.06 | 0.1957 | 0.0001584 |
| train_time | 1 | 66 | 6.55 | 0.06639 | 0.001005 | 0.7975 |